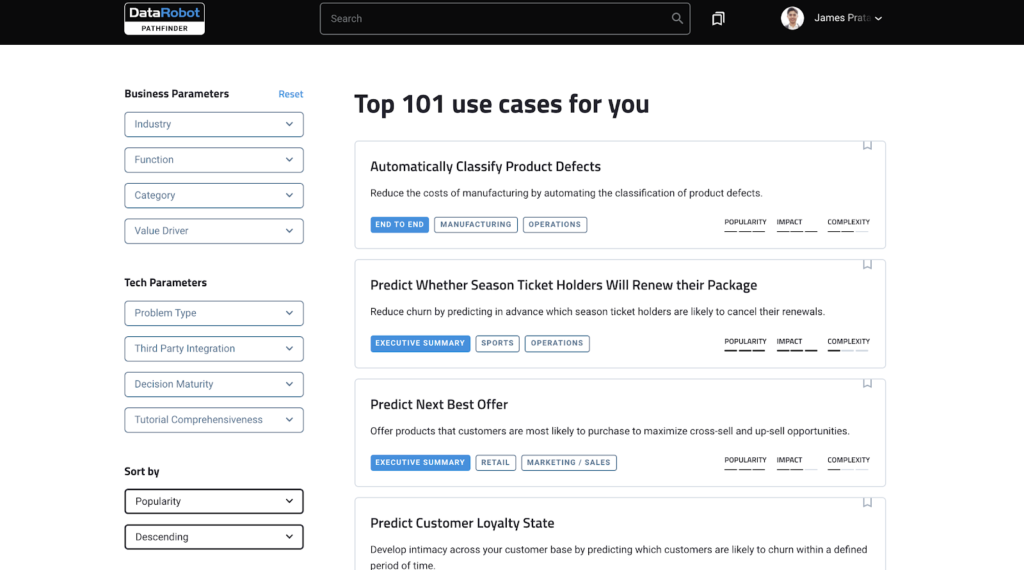
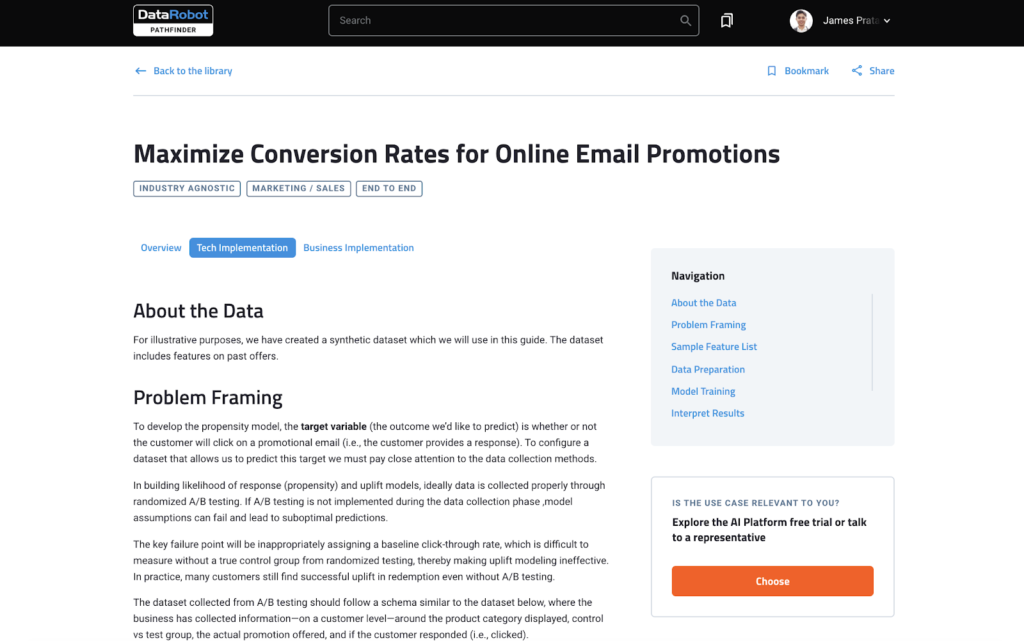
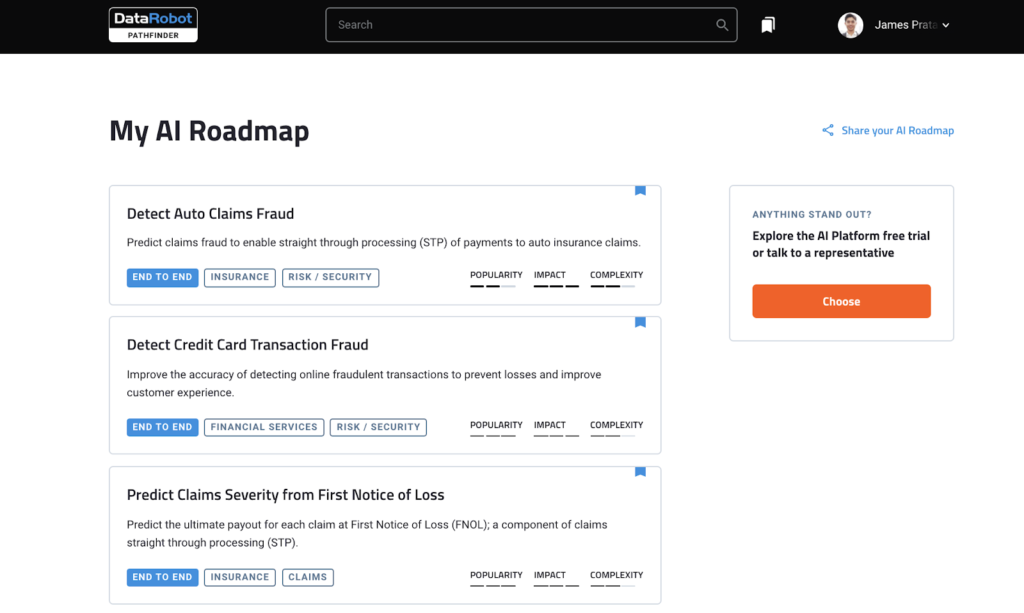
dplyr, R package that is at core of tidyverse suite of packages, provides a great set of tools to manipulate datasets in the tabular form. dplyr has a set of useful functions for “data munging”, including select(), mutate(), summarise(), and arrange() and filter().
And in this tidyverse tutorial, we will learn how to use dplyr’s filter() function to select or filter rows from a data frame with multiple examples. First, we will start with how to select rows of a dataframe based on a value of a single column or variable. And then we will learn how select rows of a dataframe using values from multiple variables or columns.
Let us get started by loading tidyverse, suite of R packges from RStudio.
library("tidyverse")
We will load Penguins data directly from cmdlinetips.com‘s github page.
path2data <- "https://raw.githubusercontent.com/cmdlinetips/data/master/palmer_penguins.csv" penguins<- readr::read_csv(path2data)
Penguins data look like this
head(penguins) ## # A tibble: 6 x 7 ## species island bill_length_mm bill_depth_mm flipper_length_… body_mass_g sex ## <chr> <chr> <dbl> <dbl> <dbl> <dbl> <chr> ## 1 Adelie Torge… 39.1 18.7 181 3750 male ## 2 Adelie Torge… 39.5 17.4 186 3800 fema… ## 3 Adelie Torge… 40.3 18 195 3250 fema… ## 4 Adelie Torge… NA NA NA NA <NA> ## 5 Adelie Torge… 36.7 19.3 193 3450 fema… ## 6 Adelie Torge… 39.3 20.6 190 3650 male
Let us subset Penguins data by filtering rows based on one or more conditions.
How to filter rows based on values of a single column in R?
Let us learn how to filter data frame based on a value of a single column. In this example, we want to subset the data such that we select rows whose “sex” column value is “fename”.
penguins %>% filter(sex=="female")
This gives us a new dataframe , a tibble, containing rows with sex column value “female”column.
## # A tibble: 165 x 7 ## species island bill_length_mm bill_depth_mm flipper_length_… body_mass_g ## <chr> <chr> <dbl> <dbl> <dbl> <dbl> ## 1 Adelie Torge… 39.5 17.4 186 3800 ## 2 Adelie Torge… 40.3 18 195 3250 ## 3 Adelie Torge… 36.7 19.3 193 3450 ## 4 Adelie Torge… 38.9 17.8 181 3625 ## 5 Adelie Torge… 41.1 17.6 182 3200 ## 6 Adelie Torge… 36.6 17.8 185 3700 ## 7 Adelie Torge… 38.7 19 195 3450 ## 8 Adelie Torge… 34.4 18.4 184 3325 ## 9 Adelie Biscoe 37.8 18.3 174 3400 ## 10 Adelie Biscoe 35.9 19.2 189 3800 ## # … with 155 more rows, and 1 more variable: sex <chr>
In our first example using filter() function in dplyr, we used the pipe operator “%>%” while using filter() function to select rows. Like other dplyr functions, we can also use filter() function without the pipe operator as shown below.
filter(penguins, sex=="female")
And we will get the same results as shown above.
In the above example, we selected rows of a dataframe by checking equality of variable’s value. We can also use filter to select rows by checking for inequality, greater or less (equal) than a variable’s value.
Let us see an example of filtering rows when a column’s value is not equal to “something”. In the example below, we filter dataframe whose species column values are not “Adelie”.
penguins %>% filter(species != "Adelie")
We now get a filtered dataframe with species other than “Adelie”
## # A tibble: 192 x 7 ## species island bill_length_mm bill_depth_mm flipper_length_… body_mass_g ## <chr> <chr> <dbl> <dbl> <dbl> <dbl> ## 1 Gentoo Biscoe 46.1 13.2 211 4500 ## 2 Gentoo Biscoe 50 16.3 230 5700 ## 3 Gentoo Biscoe 48.7 14.1 210 4450 ## 4 Gentoo Biscoe 50 15.2 218 5700 ## 5 Gentoo Biscoe 47.6 14.5 215 5400 ## 6 Gentoo Biscoe 46.5 13.5 210 4550 ## 7 Gentoo Biscoe 45.4 14.6 211 4800 ## 8 Gentoo Biscoe 46.7 15.3 219 5200 ## 9 Gentoo Biscoe 43.3 13.4 209 4400 ## 10 Gentoo Biscoe 46.8 15.4 215 5150 ## # … with 182 more rows, and 1 more variable: sex <chr>
dplyr filter() with greater than condition
When the column of interest is a numerical, we can select rows by using greater than condition. Let us see an example of filtering rows when a column’s value is greater than some specific value.
In the example below, we filter dataframe such that we select rows with body mass is greater than 6000 to see the heaviest penguins.
# filter variable greater than a value penguins %>% filter(body_mass_g> 6000)
After filtering for body mass, we get just two rows that satisfy body mass condition we provided.
# # A tibble: 2 x 7 ## species island bill_length_mm bill_depth_mm flipper_length_… body_mass_g sex ## <chr> <chr> <dbl> <dbl> <dbl> <dbl> <chr> ## 1 Gentoo Biscoe 49.2 15.2 221 6300 male ## 2 Gentoo Biscoe 59.6 17 230 6050 male
Similarly, we can select or filter rows when a column’s value is less than some specific value.
dplyr filter() with less than condition
Similarly, we can also filter rows of a dataframe with less than condition. In this example below, we select rows whose flipper length column is less than 175.
# filter variable less than a value penguins %>% filter(flipper_length_mm <175)
Here we get a new tibble with just rows satisfying our condition.
## # A tibble: 2 x 7 ## species island bill_length_mm bill_depth_mm flipper_length_… body_mass_g sex ## <chr> <chr> <dbl> <dbl> <dbl> <dbl> <chr> ## 1 Adelie Biscoe 37.8 18.3 174 3400 fema… ## 2 Adelie Biscoe 37.9 18.6 172 3150 fema…
How to Filter Rows of a dataframe using two conditions?
With dplyr’s filter() function, we can also specify more than one conditions. In the example below, we have two conditions inside filter() function, one specifies flipper length greater than 220 and second condition for sex column.
# 2.6.1 Boolean AND penguins %>% filter(flipper_length_mm >220 & sex=="female")
## # A tibble: 1 x 7 ## species island bill_length_mm bill_depth_mm flipper_length_… body_mass_g sex ## <chr> <chr> <dbl> <dbl> <dbl> <dbl> <chr> ## 1 Gentoo Biscoe 46.9 14.6 222 4875 fema…
dplyr’s filter() function with Boolean OR
We can filter dataframe for rows satisfying one of the two conditions using Boolean OR. In this example, we select rows whose flipper length value is greater than 220 or bill depth is less than 10.
penguins %>% filter(flipper_length_mm >220 | bill_depth_mm < 10)
## # A tibble: 35 x 7 ## species island bill_length_mm bill_depth_mm flipper_length_… body_mass_g ## <chr> <chr> <dbl> <dbl> <dbl> <dbl> ## 1 Gentoo Biscoe 50 16.3 230 5700 ## 2 Gentoo Biscoe 49.2 15.2 221 6300 ## 3 Gentoo Biscoe 48.7 15.1 222 5350 ## 4 Gentoo Biscoe 47.3 15.3 222 5250 ## 5 Gentoo Biscoe 59.6 17 230 6050 ## 6 Gentoo Biscoe 49.6 16 225 5700 ## 7 Gentoo Biscoe 50.5 15.9 222 5550 ## 8 Gentoo Biscoe 50.5 15.9 225 5400 ## 9 Gentoo Biscoe 50.1 15 225 5000 ## 10 Gentoo Biscoe 50.4 15.3 224 5550 ## # … with 25 more rows, and 1 more variable: sex <chr>
Select rows with missing value in a column
Often one might want to filter for or filter out rows if one of the columns have missing values. With is.na() on the column of interest, we can select rows based on a specific column value is missing.
In this example, we select rows or filter rows with bill length column with missing values.
penguins %>% filter(is.na(bill_length_mm))
In this dataset, there are only two rows with missing values in bill length column.
## # A tibble: 2 x 8 ## species island bill_length_mm bill_depth_mm flipper_length_… body_mass_g sex ## <fct> <fct> <dbl> <dbl> <int> <int> <fct> ## 1 Adelie Torge… NA NA NA NA <NA> ## 2 Gentoo Biscoe NA NA NA NA <NA> ## # … with 1 more variable: year <int>
We can also use negation symbol “!” to reverse the selection. In this example, we select rows with no missing values for sex column.
penguins %>% filter(!is.na(sex))
Note that this filtering will keep rows with other column values with missing values.
## # A tibble: 333 x 7 ## species island bill_length_mm bill_depth_mm flipper_length_… body_mass_g ## <chr> <chr> <dbl> <dbl> <dbl> <dbl> ## 1 Adelie Torge… 39.1 18.7 181 3750 ## 2 Adelie Torge… 39.5 17.4 186 3800 ## 3 Adelie Torge… 40.3 18 195 3250 ## 4 Adelie Torge… 36.7 19.3 193 3450 ## 5 Adelie Torge… 39.3 20.6 190 3650 ## 6 Adelie Torge… 38.9 17.8 181 3625 ## 7 Adelie Torge… 39.2 19.6 195 4675 ## 8 Adelie Torge… 41.1 17.6 182 3200 ## 9 Adelie Torge… 38.6 21.2 191 3800 ## 10 Adelie Torge… 34.6 21.1 198 4400 ## # … with 323 more rows, and 1 more variable: sex <chr>
The post dplyr filter(): Filter/Select Rows based on conditions appeared first on Python and R Tips.
from Python and R Tips https://ift.tt/3ld5Ht4
via Gabe's MusingsGabe's Musings